Projects
Current focus: Orbital-free density functional theory
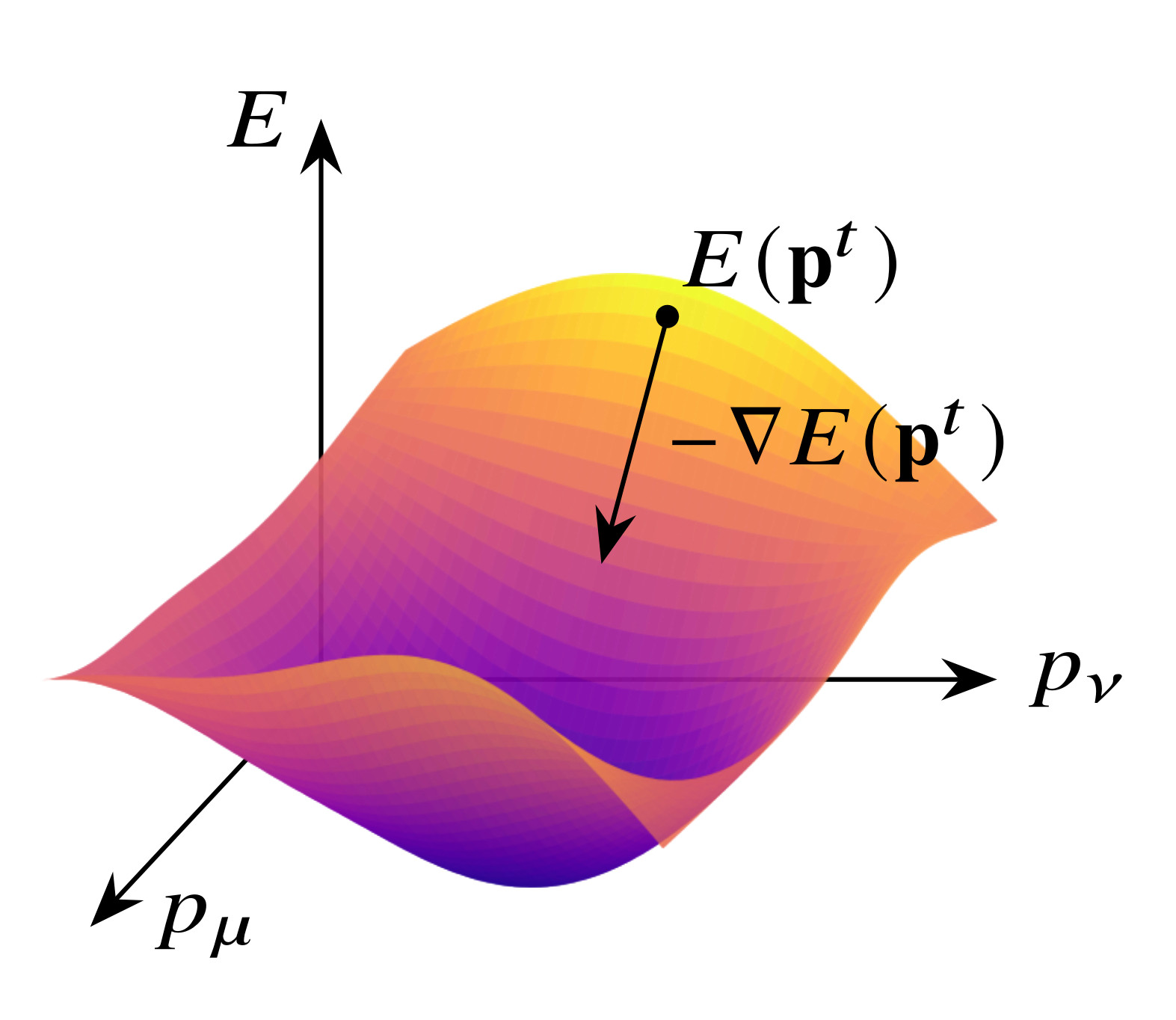
What is the kinetic energy of a given molecular ground state electron density? While the answer matters, and is known to exist in principle, so far no-one knows how to find it.
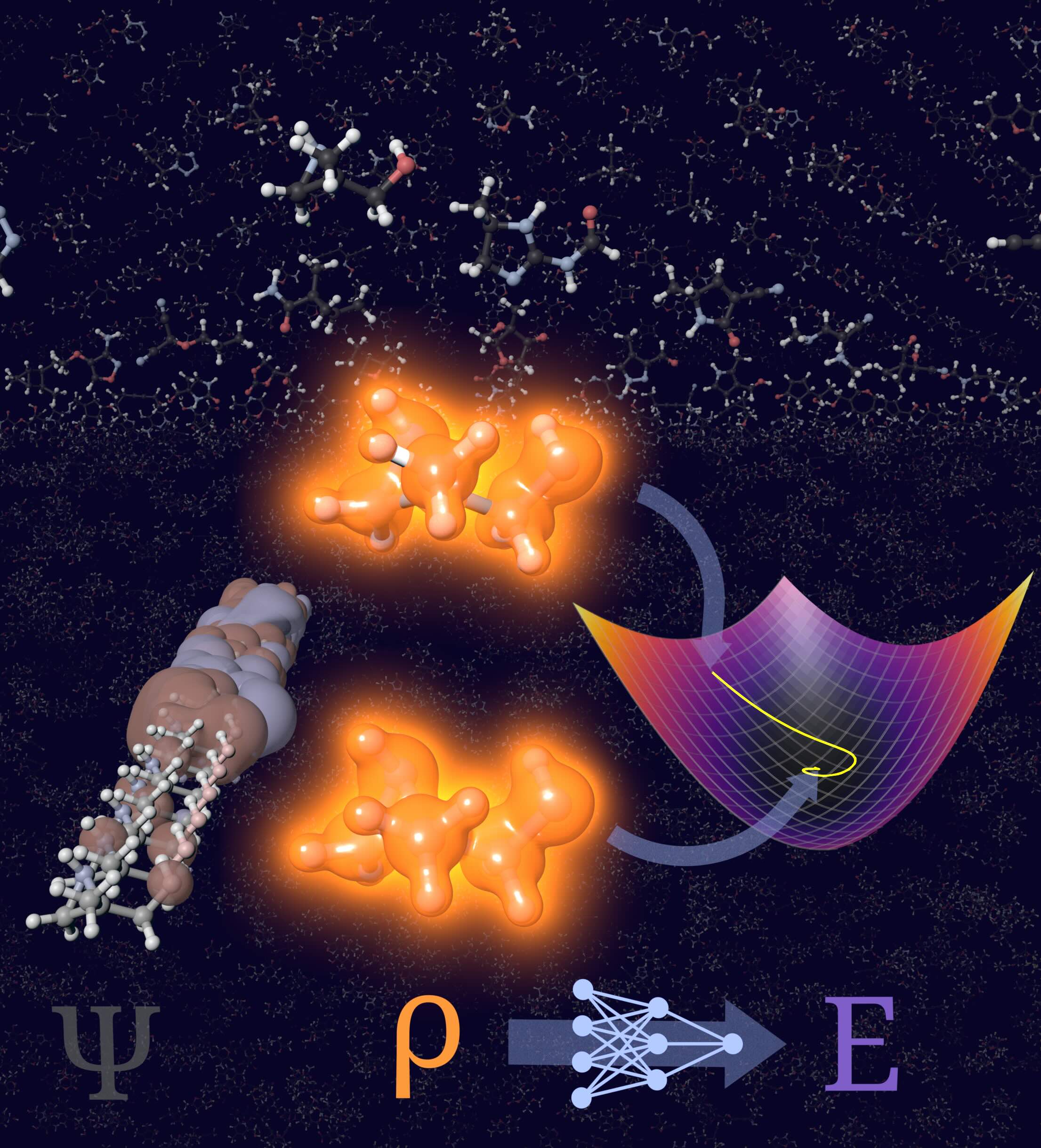
When predicting the properties of a molecule, it is natural to invoke quantum mechanics to describe the interacting electrons in terms of their many-body wave function $\psi(x_1, …, x_N)$. For sizeable molecules, this is too expensive and instead Kohn-Sham density functional theory (KS-DFT) is routinely evoked. The latter models the same system merely via one-body functions $\phi_1 (x_1)$, …, $\phi_N (x_N)$ describing non-interacting electrons moving in an effective potential created by all others. Although highly successful in practice, Kohn-Sham DFT remains a sort of band-aid solution. Why?
In the 1960s, Hohenberg and Kohn made a tantalizing discovery: for systems in their electronic ground state, knowledge of the electron density alone suffices to infer the exact electronic ground state energy, and wave functions or “orbitals” are not needed! To find the ground state electron density, one minimizes its overall energy in the potential created by atomic nuclei. The overall energy has, of course, a kinetic contribution. Vexingly, it has so far been impossible to identify the functional $T[\rho]$ yielding the exact kinetic energy of a given ground state electron density $\rho$. Finding this functional is, on the one hand, of great theoretical interest. At the same time, it paves the way for optimizing the ground state electron density without taking recourse to the much more complicated many-body wave functions. This sixty-year old promise is called “pure” or “orbital-free” DFT.
The entire team is now focused on developing machine learning methods to learn the elusive functional. If successful, this will dramatically speed up the prediction of molecular properties, paving the way to address previously inaccessible questions in the molecular and life sciences.
Selected contributions

Now headed by Anna Kreshuk at EMBL, ilastik facilitates quantitative image analysis and has many users in biology.
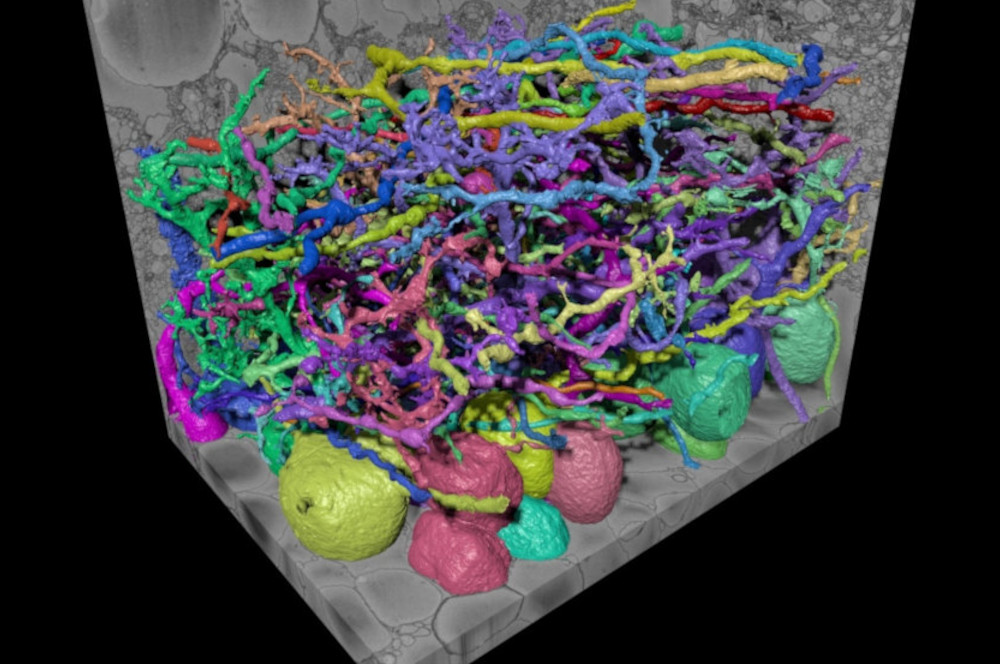
One decade of work from exactly solving the multicut problem, to the fast mutex watershed and the generalized gasp heuristic.

PlantSeg is a deep learning tool for cell instance segmentation tailored to the needs of plant biologists.